Valle-Oganov descriptor
Implementation of the fingerprint descriptor by Valle and Oganov [1] for \(k=2\) and \(k=3\) [2]. Note that the descriptor is implemented as a subclass of MBTR and can also can also be accessed in MBTR by setting the right parameters: see the MBTR tutorial.
It is advisable to first check out the MBTR tutorial to understand the basics of the MBTR framework. The differences compared to MBTR are:
The \(k=1\) term has been removed.
A radial cutoff distance r_cut is used.
In the grid setup, min is always 0 and max has the same value as r_cut.
Normalization and weighting are automatically set to their Valle-Oganov options.
periodic is set to True: Valle-Oganov normalization doesn’t support non-periodic systems.
See below for how to set k2 and k3.
Parameters species, flatten and sparse work similarly to MBTR.
Setup
Instantiating a Valle-Oganov descriptor can be done as follows:
import numpy as np
from dscribe.descriptors import ValleOganov
# Setup
vo = ValleOganov(
species=["H", "O"],
k2={
"sigma": 10**(-0.5),
"n": 100,
"r_cut": 5
},
k3={
"sigma": 10**(-0.5),
"n": 100,
"r_cut": 5
},
)
The arguments have the following effect:
In some publications, a grid parameter \(\Delta\), which signifies the width of the spacing, is used instead of n. However, here n is used in order to keep consistent with MBTR. The correlation between n and \(\Delta\) is \(n=(max-min)/\Delta+1=(r_{cutoff})/\Delta+1\).
Creation
After the descriptor has been set up, it may be used on atomic structures with the
create()-method.
from ase import Atoms
import math
water = Atoms(
cell=[[5.0, 0.0, 0.0], [0.0, 5.0, 0.0], [0.0, 0.0, 5.0]],
positions=[
[0, 0, 0],
[0.95, 0, 0],
[
0.95 * (1 + math.cos(76 / 180 * math.pi)),
0.95 * math.sin(76 / 180 * math.pi),
0.0,
],
],
symbols=["H", "O", "H"],
)
# Create ValleOganov output for the system
vo_water = vo.create(water)
print(vo_water)
print(vo_water.shape)
The call syntax for the create-function is as follows:
The output will in this case be a numpy array with shape [#positions,
#features]. The number of features may be requested beforehand with the
get_number_of_features()-method.
Examples
The following examples demonstrate some use cases for the descriptor. These examples are also available in dscribe/examples/valleoganov.py.
Visualization
The following snippet demonstrates how the output for \(k=2\) can be visualized with matplotlib. Visualization works very similarly to MBTR.
import matplotlib.pyplot as plt
import ase.data
from ase.build import bulk
# The ValleOganov-object is configured with flatten=False so that we can easily
# visualize the different terms.
nacl = bulk("NaCl", "rocksalt", a=5.64)
vo = ValleOganov(
species=["Na", "Cl"],
k2={
"n": 200,
"sigma": 10**(-0.625),
"r_cut": 10
},
flatten=False,
sparse=False
)
vo_nacl = vo.create(nacl)
# Create the mapping between an index in the output and the corresponding
# chemical symbol
n_elements = len(vo.species)
imap = vo.index_to_atomic_number
x = np.linspace(0, 10, 200)
smap = {index: ase.data.chemical_symbols[number] for index, number in imap.items()}
# Plot k=2
fig, ax = plt.subplots()
legend = []
for i in range(n_elements):
for j in range(n_elements):
if j >= i:
plt.plot(x, vo_nacl["k2"][i, j, :])
legend.append(f'{smap[i]}-{smap[j]}')
ax.set_xlabel("Distance (angstrom)")
plt.legend(legend)
plt.show()
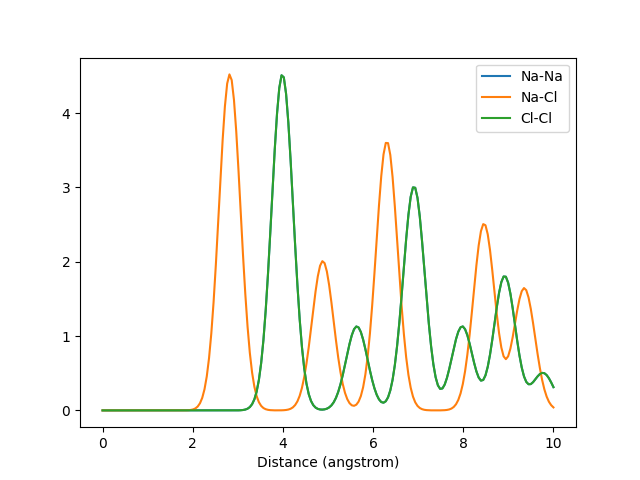
The Valle-Oganov output for k=2. The graphs for Na-Na and Cl-Cl overlap due to their identical arrangement in the crystal.
Setup with MBTR class
For comparison, the setup for Valle-Oganov descriptor for the previous structure, but using the MBTR class, would look like the following.
from dscribe.descriptors import MBTR
mbtr = MBTR(
species=["Na", "Cl"],
k2={
"geometry": {"function": "distance"},
"grid": {"min": 0, "max": 10, "sigma": 10**(-0.625), "n": 200},
"weighting": {"function": "inverse_square", "r_cut": 10},
},
normalize_gaussians=True,
normalization="valle_oganov",
periodic=True,
flatten=False,
sparse=False
)
Side by side with MBTR output
A graph of the output for the same structure created with different descriptors. Also demonstrates how the Valle-Oganov normalization for k2 term converges at 1.
nacl = bulk("NaCl", "rocksalt", a=5.64)
decay = 0.5
mbtr = MBTR(
species=["Na", "Cl"],
k2={
"geometry": {"function": "distance"},
"grid": {"min": 0, "max": 20, "sigma": 10**(-0.625), "n": 200},
"weighting": {"function": "exp", "scale": decay, "threshold": 1e-3},
},
periodic=True,
flatten=False,
sparse=False
)
vo = ValleOganov(
species=["Na", "Cl"],
k2={
"n": 200,
"sigma": 10**(-0.625),
"r_cut": 20
},
flatten=False,
sparse=False
)
mbtr_output = mbtr.create(nacl)
vo_output = vo.create(nacl)
n_elements = len(vo.species)
imap = vo.index_to_atomic_number
x = np.linspace(0, 20, 200)
smap = {index: ase.data.chemical_symbols[number] for index, number in imap.items()}
fig, ax = plt.subplots()
legend = []
for key, output in {"MBTR": mbtr_output, "Valle-Oganov": vo_output}.items():
plt.plot(x, output["k2"][0, 1, :])
legend.append(f'{key}: {smap[0]}-{smap[1]}')
ax.set_xlabel("Distance (angstrom)")
plt.legend(legend)
plt.show()
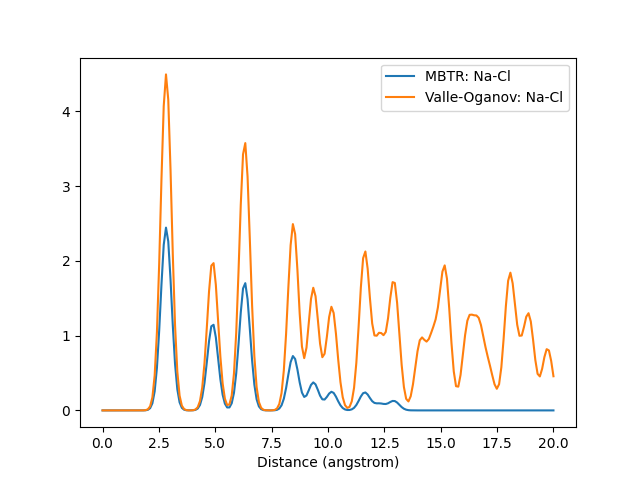
Outputs for k=2. For the sake of clarity, the cutoff distance has been lengthened and only the Na-Cl pair has been plotted.
- 1
Mario Valle and Artem R Oganov. Crystal fingerprint space–a novel paradigm for studying crystal-structure sets. Acta Crystallographica Section A: Foundations of Crystallography, 66(5):507–517, 2010.
- 2
Malthe K Bisbo and Bjørk Hammer. Global optimization of atomistic structure enhanced by machine learning. arXiv preprint arXiv:2012.15222, 2020.